- Retina Research Foundation
- About RRF
- Pilot Study Grants
- Grant Recipients 2025
- Samuel M. Wu, PhD
- Yingbin Fu, PhD
- Graeme Mardon, PhD
- Wei Li, PhD
- Yuqing Huo, MD, PhD
- Rui Chen, PhD
- Wenbo Zhang, PhD
- Curtis Brandt, PhD
- Lih Kuo, PhD
- Timothy Corson, PhD
- Jianhai Du, PhD
- Francesco Giorgianni, PhD
- James Monaghan, PhD
- Seongjin Seo, PhD
- Andrius Kazlauskas, PhD
- Erika D. Eggers, PhD
- Ann C. Morris, PhD
- Ming Zhang, MD, PhD
- Christine Sorenson, PhD
- Alex J. Smith, PhD
- Jeffrey M. Gross, PhD
- David M. Wu, MD, PhD
- Kinga Bujakowska, PhD
- Eric Weh, PhD
- Ching-Kang Jason Chen, PhD
- Jakub K. Famulski, PhD
- Thanh Hoang, PhD
- Georgia Zarkada, MD, PhD
- Eleftherios Paschalis Ilios, PhD
- Oleg Alekseev, MD, PhD
- Erika Tatiana Camacho, PhD
- Patricia R. Taylor, PhD
- Elizabeth Vargis, PhD
- Publications
- Grant Guidelines and Information
- Grant Application
- Grant Recipients 2025
- Research Programs
- Contact Us
- Giving
- RRF History
- Home
Helmerich Chair
Walter H. Helmerich Research Chair (2017 to present)
Associate Director, McPherson Eye Research Institute
Kevin W Eliceiri, PhD
Laboratory for Optical and Computational Instrumentation
Departments of Biomedical Engineering and Medical Physics
Morgridge Institute for Research
University of Wisconsin
Madison, WI
Dr. Eliceiri’s Research Project
Computational Imaging of the Cellular Microenvironment
Overview
Dr. Eliceiri’s research focuses on the development of computational and optical approaches for bioimaging. The overarching goal of the Eliceiri lab is cellular scale biomarker discovery for disease onset and progression through quantitative imaging and computational analysis. He has been conducting research on computational and optical approaches to live cell imaging since 2000 with a focus on multidimensional fluorescence approaches for cell studies and image informatics tools for image analysis. In particular, his lab is developing new approaches for dynamic cell imaging in vivo including Multiphoton Laser Scanning Microscopy (MPLSM), Second Harmonic Generation (SHG), FRET and fluorescence lifetime imaging (FLIM) approaches. A major focus of his research is to develop and apply non-invasive optical and computational approaches for understanding the role of the cellular microenvironment in normal and diseased processes. Of particular interest is the development and use of advancing imaging and image analysis approaches to study cell-cell, cell-matrix and endothelial-stromal interactions and metabolism.
Computational
Dr. Eliceiri is also actively engaged in developing quantitative approaches for analyzing multidimensional fluorescence data sets. . Much of this work has been on open source approaches for analyzing and extracting cell scale information from fluorescence 3D time-lapse (4D) datasets. From the beginning of these efforts the Eliceiri lab has focused on the open source image analysis application, ImageJ. Recently his lab launched their ImageJ2 efforts, to modernize and extend ImageJ while still offering complete backwards compatibility. These open source imaging efforts have also included leveraging modern computational and machine learning tools for extracting meaningful information from visual data in real time. For example, the Eliceiri lab recently partnered with Google on the implementation of a machine learning system within an image analysis package. This can be run during or after data acquisition to extract key features of a cell targeted by disease.
Optical
The Eliceiri lab has built a number of novel instrumentation approaches involving multiphoton laser scanning microscope, second harmonic generation imaging, spectral, lifetime and polarization to investigate the role of cell scale biological approaches. More recently they have been engaging in computational optics and multiscale imaging. In computational optics we are designing systems where computational code can be run at runtime, to allow for analysis during acquisition. These efforts revolve around the concept of “Smart Microscopy” where machine learning algorithms can be used during acquisition to help drive the result. As well the Eliceiri lab is doing real time analysis of cell-based features such as fluorescence lifetime based metabolic mapping. In multiscale imaging they are trying to track the relevance of cell based changes to tissue level changes by building a class of imaging hardware that can resolve structures from nm to mms. The Eliceiri lab is currently modifying one of their systems to include a variety of optical imaging methods of different spatial scales (SHG, MPSLM, EBS and OCT) and integrating that with quantitative ultrasound to better capture the role of cell levels changes in bulk tissue composition.
Helmerich Chair (2012 – 2017)
Medical Genetics
University of Wisconsin
Madison, WI
Dr. Ikeda’s Research Project
Identification of Genetic Factors Affecting Aging of the Retina
Current Research Interests
Dr. Ikeda uses mouse models to study the genetic and molecular mechanisms of aging. His laboratory studies a mouse mutant showing similar symptoms as observed in age-related macular degeneration (AMD) patients. He has identified the mutation in the gene (Tmem135) associated with mitochondria functions and confirmed that the mutation is indeed causing the AMD-like symptoms.
Another major project is to identify genes that determine the severity of aging symptoms in the retina including neurodegeneration, synaptic abnormality, and inflammation using two mouse strains, one of which shows retinal aging symptoms earlier than the other. He has found that a mutation in the bloom syndrome gene (Blm) gene involved in DNA damage repair is responsible for the early-onset of aging symptoms and that Blm may have a role in the mitochondrial function.
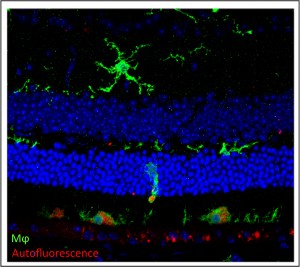
The retina of a mutant mouse with accelerated aging symptoms show AMD-like phenotypes such as accumulation of autofluorescence and inflammation.Specifically, Dr. Ikeda’s laboratory aims to identify gene mutations that lead to early onset of aging phenotypes in the mouse retina. His laboratory also aims to identify genes that cause the difference in the severity of age-dependent retinal abnormalities observed between two mouse inbred strains. Dr. Ikeda hopes that identification of these genes will allow him to elucidate the molecular mechanisms causing the age-dependent retinal abnormalities, and therefore, enhance our understanding of age-dependent retinal diseases and aging of the retina.