- Retina Research Foundation
- About RRF
- Pilot Study Grants
- Grant Recipients 2025
- Samuel M. Wu, PhD
- Yingbin Fu, PhD
- Graeme Mardon, PhD
- Wei Li, PhD
- Yuqing Huo, MD, PhD
- Rui Chen, PhD
- Wenbo Zhang, PhD
- Curtis Brandt, PhD
- Lih Kuo, PhD
- Timothy Corson, PhD
- Jianhai Du, PhD
- Francesco Giorgianni, PhD
- James Monaghan, PhD
- Seongjin Seo, PhD
- Andrius Kazlauskas, PhD
- Erika D. Eggers, PhD
- Ann C. Morris, PhD
- Ming Zhang, MD, PhD
- Christine Sorenson, PhD
- Alex J. Smith, PhD
- Jeffrey M. Gross, PhD
- David M. Wu, MD, PhD
- Kinga Bujakowska, PhD
- Eric Weh, PhD
- Ching-Kang Jason Chen, PhD
- Jakub K. Famulski, PhD
- Thanh Hoang, PhD
- Georgia Zarkada, MD, PhD
- Eleftherios Paschalis Ilios, PhD
- Oleg Alekseev, MD, PhD
- Erika Tatiana Camacho, PhD
- Patricia R. Taylor, PhD
- Elizabeth Vargis, PhD
- Publications
- Grant Guidelines and Information
- Grant Application
- Grant Recipients 2025
- Research Programs
- Contact Us
- Giving
- RRF History
- Home
Rui Chen, PhD
 Gavin Herbert Eye Institute – Center for Translational Vision Research, Department of Ophthalmology
Gavin Herbert Eye Institute – Center for Translational Vision Research, Department of Ophthalmology
University of California, Irving
Irvine, CA
EMMETT A. HUMBLE RESEARCH PROJECT
Identification and Functional Analysis of Genes Involved in Retinal Diseases and DevelopmentCurrent Research Interests
The long-term goal of Dr. Chen’s project is to improve the ability to prevent, diagnose, and treat human retinal diseases. Since many human eye diseases genes are involved in normal eye development, Dr. Chen’s lab uses animal model systems to study retinal development. His research interests are to identify and conduct functional characterization of novel disease genes underlying human retinal disorders. Results obtained from studies can be directly translated into our understanding and form the basis of developing optimal treatment of human eye diseases. To learn more about Dr. Chen’s groundbreaking work, follow this link to Dr. Chen’s Lab
Plans for 2025
During 2025, Dr. Chen will continue his efforts to characterize the newly identified retinal disease gene DDX41, whose function in the retina has not previously been investigated. Dr. Chen has recently established a mutant mice model for Ddx41 to study the gene’s function, which exhibits retinal degeneration mimicking the patient phenotype. He will perform a detailed phenotype characterization to provide critical insights of DDX41 pathophysiology in patients. The lab will also test the feasibility of performing gene therapy to rescue the photoreceptor degeneration phenotype due to mutation in DDX41 using the mice model. Finally, Dr. Chen plans to continue performing sequencing analysis and follow up functional validation and studies of novel candidate pathogenic variants and disease genes will be performed.
Progress in 2024
Excellent progress was made in the investigation of the function of Tlcd3b, a novel inherited retinal disease (IRD) gene, in the retina. In the profiling of ceramide in the Tcld3b mutant retina, Dr. Chen observed that, as expected, the level of C16-C20 ceramide is significantly reduced compared to control, and he published his findings in Genetics in Medicine. In addition, the lab tested whether the Tlcd3b mutant phenotype in the retina can be rescued by overexpression of canonical CerS, and it appears that Tlcd3b retina phenotype can be partially rescued by CerS with the best rescue observed for CerS5. The team continues to investigate the phenotype and interaction between CerS with Tlcd3b, and published a manuscript describing these results in Disease Models and Mechanisms.
As published in IOVS, Dr. Chen performed gene therapy on the Tlcd3b KO model and demonstrated significant rescue via gene replacement therapy. This pre-clinical result lays the foundation for future development of human gene therapy.
Dr. Chen continues to perform his functional studies for candidate disease genes and gene therapy. By analyzing whole genome sequencing data on patients with inherited retinal diseases, he continues to identify novel candidate disease genes, including DDX41, whose function in the retina has not been investigated and will the focus of his research in 2025.
Progress in 2023
Dr. Chen’s laboratory team had previously completed the panel sequencing for nearly 1000 LCA patients from other collaborators. Among them, approximately 750 patients received positive molecular diagnosis while the remaining 250 patient remain unsolved. In 2023, the lab began whole genome sequencing (WGS). Analysis of the WGS allows for the identification of novel mutations that were previously missed by the panel and whole exome sequencing technologies. results of this effort led to publication in Hum Mol Genet and Genes.
In parallel, the team performed follow-up functional studies of TLCD3B, a novel gene associated with IRD identified in our previous funding period. They also performed profiling of ceramide in the Tcld3b mutant retina and observed that, as expected, the level of C16-C20 ceramide is significantly reduced compared to control. These findings were published in Genetic in Medicine.
To better understand the potential interaction between canonical CerS and Tlcd3b, Dr. Chen tested whether Tlcd3b mutant phenotype in the retina can be rescued by overexpression of canonical CerS. Based on current data, Tlcd3b retina phenotype can be partially rescued by CerS with the best rescue observed for CerS5. The lab further investigated the phenotype and interaction between CerS with Tlcd3b. A manuscript describing these results has now been published at Dis Model Mech. Progress throughout the year led to four publications.
Progress in 2022
The goal of Dr. Chen’s project prior to this year has been specifically to identify novel genes involved in Leber congenital amaurosis (LCA), to perform the necessary associated disease mechanism studies, and to develop novel therapeutic approaches to treat the disease. To date, a total of 256 genes have been identified that are associated with heritable human eye diseases, indicating the high heterogeneity of the disease. Therefore, an understanding of molecular mechanisms of retina disease is an essential first step toward designing personalized treatments for inherited eye diseases. As one of the most common hereditary causes of visual impairment in infants and children, LCA accounts for more than 5% of all retinal dystrophies (Koenekoop, 2004; Perrault et al., 1996). Due to it genetics heterogeneity, accurate molecular diagnosis of LCA patients and administration of appropriate interventions is essential for treatment. However, despite its heterogeneity, LCA shares many common molecular mechanisms with other retinal dystrophies, such as Retinitis Pigmentosa (RP) and Bardet-Biedl Syndrome (BBS). Therefore, by studying LCA, valuable insights or understanding of other retinal degenerative dystrophies will also be learned. Subsequent functional analysis of the novel disease genes identified in this study through modeling in organisms is the first step toward both early diagnosis of at-risk individuals at risk as well as developing reagents for treatment of retinal disease, including gene and drug therapy.
Dr. Chen’s team collaboratively had previously completed panel sequencing for approximately 1000 LCA patients. Of these patients, about 750 received a positive molecular diagnosis while the remaining 250 patient remain unsolved. In 2022, Dr. Chen began performing whole genome sequencing (WGS). Analysis of the WGS allows for identification of novel mutations that were previously missed by panel and whole exome sequencing technologies. One type of mutations is deep intronic cryptic splicing mutations, research findings that were published in Frontiers in Genetics. Dr. Chen established a mutant mice model for Tlcd3b, a novel human retinal disease gene identified by his laboratory. Using this mice model, in the past year, he tested the feasibility of performing gene therapy to rescue the photoreceptor degeneration phenotype. His results demonstrated that gene replacement therapy is effective in treating the Tlcd3b mutant mice, laying the foundation for future human therapy development. The manuscript describing these findings was published in IVOS, and overall progress during the year led to four publications.
Progress in 2021
Dr. Chen and his laboratory team made significant progress in both areas of focus. They completed the panel sequencing for all Saudi patient cohort, Iran cohort, as well the 600 LCA patients from other collaborators. In addition, for all samples without a good candidate, the whole exome sequencing was completed, totaling over 300 patients. Dr. Chen upgraded the data to whole genome sequencing for 50 familiar patient samples. In 2021, Dr. Chen published the identification of novel disease gene TLCD3B in Genetics in Medicine. In addition, his team completed the gene therapy study focusing on distinguishing the function of the two isoforms of REEP6, a disease gene Dr. Chen discovered previously. The manuscript describing the work was published in Molecular Human Genetics. Finally, animal models were generated for CWC27 and TLCD3B newly, for which functional studies and developing novel therapeutic approach are currently underway. In total, research progress led to five publications in 2021.
Progress in 2020
Dr. Chen made significant progress on both aims in 2020. His laboratory identified multiple LCA and RP disease genes, including IFT81, REEP6, CWC27, and most recently, FAM57B. Based on these discoveries, his team will continue their efforts to identify novel LCA disease genes and follow up with functional studies that are critical for understanding the mechanisms of the disease. Animal models have been generated for some of these newly discovered disease genes, and functional studies and developing novel therapeutic approach are currently underway. Dr. Chen’s important progress led to four
Progress in 2019
In 2019, exceptional progress was achieved in each specific aim of Dr. Chen’s research. Dr. Chen focused on identification of additional genes whose mutations cause Leber congenital amaurosis (LCA), the most common hereditary cause of visual impairment in infants and children. It has been estimated that mutations in known LCA genes account for about 70% of all cases. To clone additional LCA disease genes, his research team collected DNA samples from 38 consanguineous families with recessive LCA as well as 800 sporadic cases. Exome and whole genome sequencing will be performed for these samples to identify novel disease genes, which will assist the development of new diagnostic tools and treatments (Aim 1). In addition, since mutations in LCA disease genes also cause other retinal dystrophies, isolation of additional LCA disease genes will provide important insights into the molecular mechanisms underlying LCA and other retinal dystrophies.
In parallel, Dr. Chen pursued development of potential therapeutic approaches using the disease animal models established in his laboratory. To further illustrate gene functions, his lab conducted functional analysis of identified novel disease genes necessary to retina function (Aim 2). Characterization of the function of this important gene subfamily provides the foundation for creating new tools for early diagnosis of at-risk individuals and developing reagents for treatment of retinal disease, including gene therapy.
Progress in 2018
In 2018, efforts were continued in novel LCA disease gene identification and follow up functional studies, which remain critical to understanding of the mechanisms of the disease.
Specific Aims: Aim 1. Identify Novel LCA Disease Genes; Aim 2. Functional analysis of candidate disease genes
Dr. Chen’s research team made significant progress in both aims. In brief, the team identified multiple LCA and RP disease genes, including IFT81, REEP6, and CWC27. Building on the success, they continued efforts in novel LCA disease gene identification and follow up functional studies, both critical to understanding the mechanism of the disease. In addition, animal models were generated for some of the newly discovered disease genes, for which functional studies and novel therapeutic approaches were conducted.
Progress in 2017
The application for RRF funding in 2017 contained two Specific Aims. Excellent progress was made on each Aim and is summarized below:
Aim 1. Identify Novel LCA Disease Genes – Dr. Chen’s laboratory completed the panel sequencing for all Saudi patient cohort, Iran cohort, as well the 600 LCA patients from other collaborators. Initial analysis was completed for these patients and several candidate novel disease genes were identified.
Aim 2. Functional analysis of candidate disease genes – Dr. Chen’s laboratory established and began characterizing mouse models for novel disease genes, REEP6 and CWC27, to allow for additional mechanistic studies and development of novel therapeutic methods for the disease.
Progress in 2016
Dr. Chen completed the sequencing for all Saudi patient cohort as well as the 600 LCA patients from other collaborators ans was sequencing patients from Iran and the newly recruited 300 samples. Through exome sequencing, Dr. Chen identified two novel LCA and RP disease genes in 2016, including CEP78 and RCBTB2. In addition, animal models were generated for some of the newly discovered disease genes, for which functional studies were undertaken. The project benefited patients in several ways, such as when positive results were obtained, the molecular mutation screen result provided critical information for diagnosis, prognosis, and potential treatment.
Progress in 2015
Research milestones in 2015 included: 1) Identification of novel retina disease gene – Dr. Chen completed the sequencing for all Saudi patient cohort as well as the 600 LCA patients from other collaborators. Initial analysis was performed for the patients, and several candidate novel disease genes were identified. 2) Functional analysis of candidate disease genes – Through exome sequencing, Dr. Chen identified several novel LCA and RP disease genes, such as ATF6 and CLAUP1. He performed functional studies of these novel disease genes using both cell and animal model. Dr. Chen’s lab generated a 2mouse model for these disease genes, which allowed for additional mechanistic studies and development of a therapeutic method of the disease.
Progress in 2014
Dr. Chen’s project identifies novel genes involved in human retinal disorders, conduct functional analysis, and develop therapy of these disease genes using a model organism such as Mus musculus. He completed the sequencing for a Saudi patient cohort as well as more than 400 Leber congenital amaurosis (LCA) and retinitis pigmentosa (RP) patients with various ethnic backgrounds. Through exome sequencing and functional studies, Dr. Chen identified several novel LCA and RP disease genes, such as HK1 and ATF6. This work was accepted for publication in IOVS. Further analysis of the data set continued.
Progress in 2013
Dr. Chen collected DNA samples from 38 consanguineous families with recessive Leber congenital amaurosis (LCA) as well as 800 sporadic cases in order to clone additional LCA disease genes. His laboratory applied cutting edge sequencing technology in cloning disease genes underlying LCA and performed whole exome sequencing on a large cohort of LCA patients. They identified several novel mutation and candidate novel disease genes for which validation and functional analysis was undertaken.
Progress in 2012
Dr. Chen’s laboratory successfully cloned the LCA3 disease gene. To better understand its normal function in the retina, Dr. Chen examined the expression pattern of Spata7 in the developing and mature mouse retina and found that LCA3 is expressed in multiple layers, most strongly in the inner segment of photoreceptor cells. Results suggest a novel disease mechanism of LCA in which LCA3 functions as part of the protein transporter vesicle, potentially as a cargo receptor, in photoreceptor cells function. Significant progress was made on experiments identifying additional LCA disease genes. Exome sequencing of 110 isolated LCA patients has been completed and analysis of these data is ongoing.
Dr. Chen focused on identification of additional genes whose mutations cause Leber congenital amaurosis (LCA), the most common hereditary cause of visual impairment in infants and children. To clone additional LCA disease genes, his laboratory collected DNA samples from 38 consanguineous families with recessive LCA as well as 200 sporadic cases. Exome sequencing was performed for these samples to identify novel genes, which assists the development of new diagnostic tools and treatments.
In parallel, they studied the function of the POU-domain subfamily 6 genes during normal retinal development. Characterization of the function of this important gene subfamily provided the basis toward creating new tools for both early diagnosis of individuals at work and developing reagents for treatment of retinal disease, including gene therapy.
Protein aggregation assay in cell culture for NMNAT1 mutant allele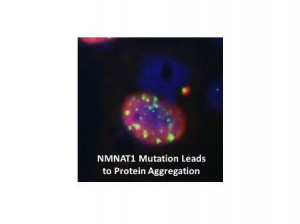
Progress in 2011
Dr. Chen’s laboratory applied the cutting edge sequencing technology in cloning disease genes underlying LCA and performed whole exome sequencing on 50 patients. Based on their preliminary results, they identified three novel LCA/RP disease genes, including CEP164, KCNJ13 and NMNAT1.
Results suggest a novel disease mechanism of LCA in which LCA3 functions as part of the protein transporter vesicle, potentially as a cargo receptor, in photoreceptor cells function. Dr. Chen made significant progress in identifying additional LCA disease genes through exome sequencing.
Leber congenital amaurosis (LCA) is one of the most common hereditary causes of visual impairment in infants and children, which accounts for more than 5% of all retinal dystrophies. The clinical phenotype of LCA can be extremely severe and it is characterized by several visual perturbations identifiable at birth or within the first year of life, including infantile nystagmus, a variety of fundus changes, and minimal or absent responses on the electroretinogram, each of which occurs with an autosomal recessive mode of inheritance.
In Dr. Chen’s project, his laboratory aimed to identify the underlying mutations for LCA, which comprised the essential first step for understanding the molecular mechanisms and designing proper treatment for this disease.
Based on homozygosity mapping of all the consanguineous LCA families with no known mutations (Li et al., 2009), Dr. Chen’s laboratory identified numerous LCA disease candidate loci. In total, they completed homozygosity mapping for 23 LCA families, each containing homozygous regions which range in size from 2 Mb to more than 200 Mb. Among them, candidate loci from five families, KKESH008, KKESH012, KKESH013, KKESH086, and KKESH205 are less than 10 Mb in size. In addition, candidate loci from two families, KKESH038 and KKESH050, are between 10-20 Mb in size. Furthermore, candidate loci from seven families are greater than 20 Mb but below 100 Mb in size. Finally, candidate loci from ten families are greater than 100 Mb in size with the largest at 243 Mb.
Whole exome capture sequencing was performed on the first 15 families. Very interestly, mutations in previous known retinal disease genes, including BBS4, MyoA, and NPHP5, were identified in these families. Since patients in this family did not display other symptom associated with mutations in these genes, these results provide new link between these syndromic retinal disease genes to non syndromic LCA, an emerging theme that had been observed concurrently by several groups. Furthermore, Dr. Chen identified several novel candidate LCA disease genes, including CEP164. Further evaluation of these novel disease genes was conducted.

